ConWhAt Atlases¶
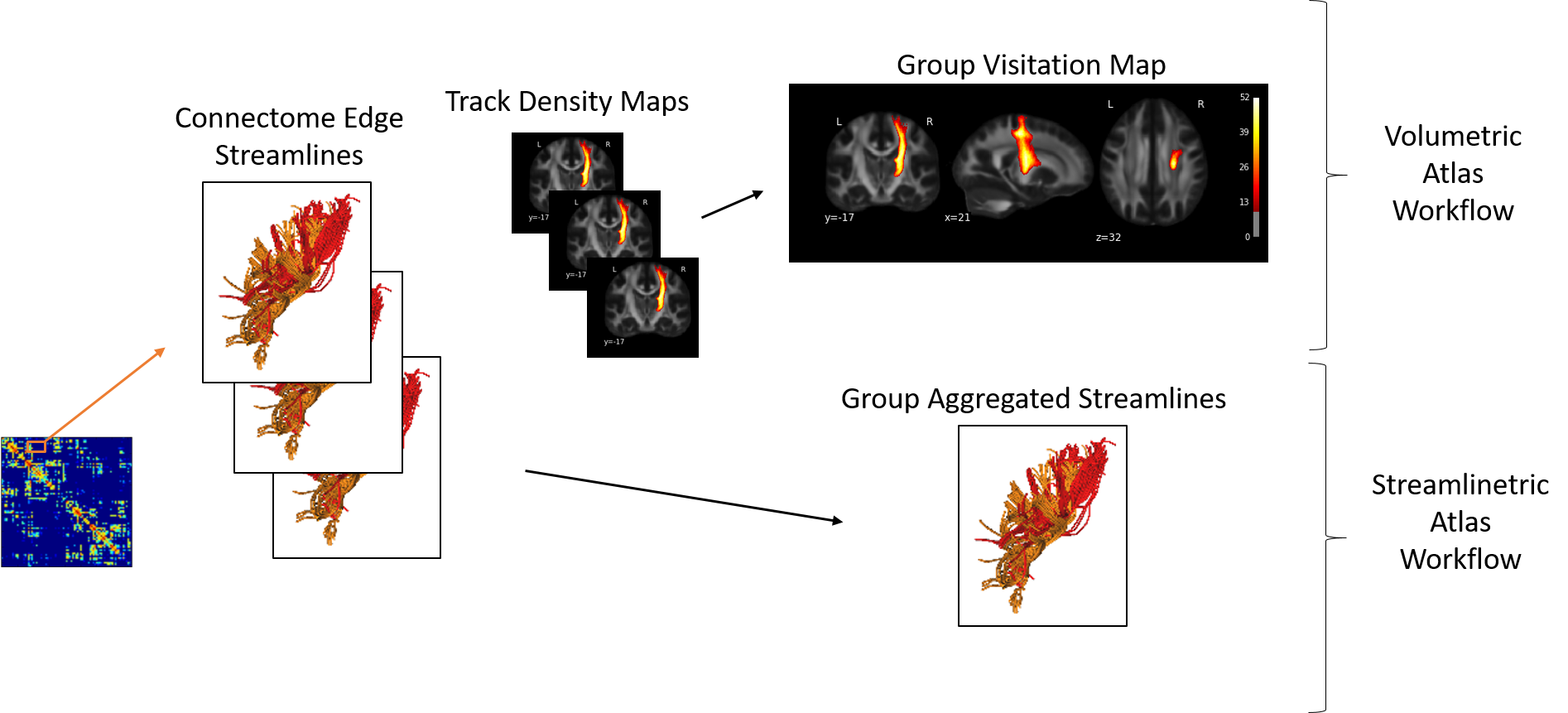
A central component of ConWhAt is the large set of connectome-based white matter atlases we have developed for use with the software. The atlas construction methodology is described in detail in Griffiths & McIntosh (in prep). Here we give a brief summary:
All of the ConWhAt atlases were constructed from on dipy deterministic whole-brain HARDI tractography reconstructions using the HCP WU-Minn corpus. Whole-brain streamline sets were segmented with region pairs using a broad set of brain parcellations, yielding anatomical connectivity matrices and connectome edge-labelled streamline sets. The streamlines are then entered into both volumetric and a streamlinetric atlas construction pipelines: