Exploring the atlases¶
There are four different atlas types in ConWhat, corresponding to the 2 ontology types (Tract-based / Connectivity-Based) and 2 representation types (Volumetric / Streamlinetric).
(More on this schema here)
>>> # ConWhAt stuff
>>> from conwhat import VolConnAtlas,StreamConnAtlas,VolTractAtlas,StreamTractAtlas
>>> from conwhat.viz.volume import plot_vol_scatter
>>> # Neuroimaging stuff
>>> import nibabel as nib
>>> from nilearn.plotting import plot_stat_map,plot_surf_roi
>>> # Viz stuff
>>> %matplotlib inline
>>> from matplotlib import pyplot as plt
>>> import seaborn as sns
>>> # Generic stuff
>>> import glob, numpy as np, pandas as pd, networkx as nx
We’ll start with the scale 33 lausanne 2008 volumetric connectivity-based atlas.
Define the atlas name and top-level directory location
>>> atlas_dir = '/scratch/hpc3230/Data/conwhat_atlases'
>>> atlas_name = 'CWL2k8Sc33Vol3d100s_v01'
Initialize the atlas class
>>> vca = VolConnAtlas(atlas_dir=atlas_dir + '/' + atlas_name,
atlas_name=atlas_name)
loading file mapping
loading vol bbox
loading connectivity
This atlas object contains various pieces of general information
>>> vca.atlas_name
'CWL2k8Sc33Vol3d100s_v01'
>>> vca.atlas_dir
'/scratch/hpc3230/Data/conwhat_atlases/CWL2k8Sc33Vol3d100s_v01'
Information about each atlas entry is contained in the vfms
attribute, which returns a pandas dataframe
Additionally, connectivity-based atlases also contain a networkx
graph object vca.Gnx, which contains information about each
connectome edge
>>> vca.Gnx.edges[(10,35)]
{'attr_dict': {'4dvolind': nan,
'fullname': 'L_paracentral_to_L_caudate',
'idx': 1637,
'name': '10_to_35',
'nii_file': 'vismap_grp_11-36_norm.nii.gz',
'nii_file_id': 1637,
'weight': 50.240000000000002,
'xmax': 92,
'xmin': 61,
'ymax': 167,
'ymin': 75,
'zmax': 92,
'zmin': 62}}
Individual atlas entry nifti images can be grabbed like so
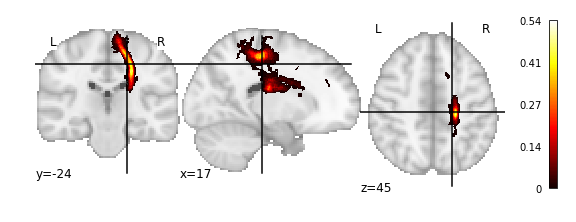
>>> img = vca.get_vol_from_vfm(1637)
getting atlas entry 1637: image file /scratch/hpc3230/Data/conwhat_atlases/CWL2k8Sc33Vol3d100s_v01/vismap_grp_11-36_norm.nii.gz
>>> plot_stat_map(img)
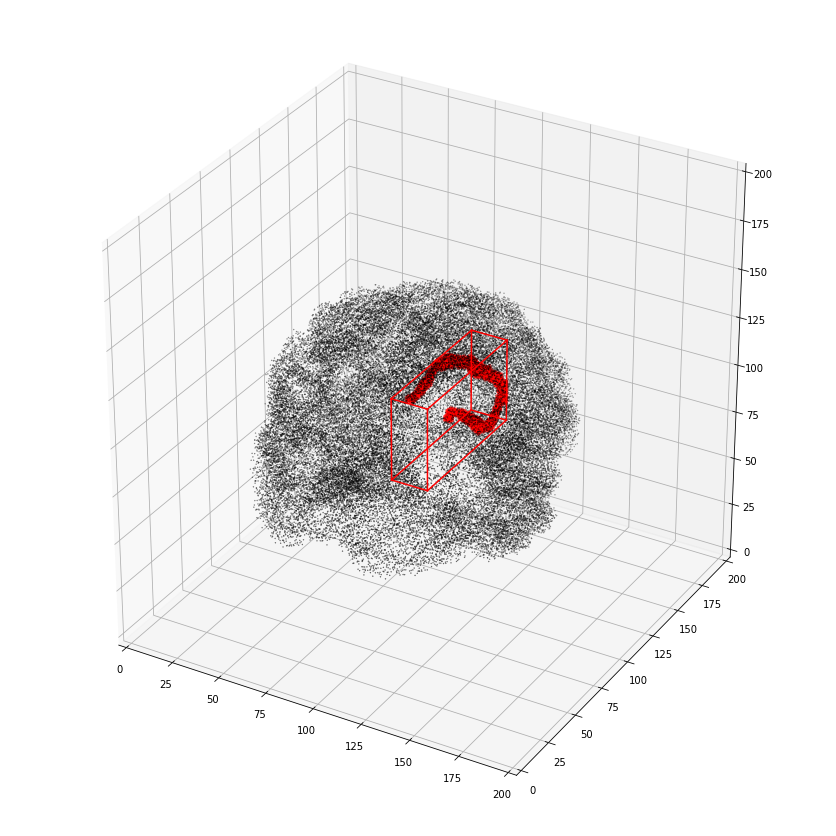
Or alternatively as a 3D scatter plot, along with the x,y,z bounding box
>>> vca.bbox.ix[1637]
xmin 61
xmax 92
ymin 75
ymax 167
zmin 62
zmax 92
Name: 1637, dtype: int64
>>> ax = plot_vol_scatter(vca.get_vol_from_vfm(1),c='r',bg_img='nilearn_destrieux',
>>> bg_params={'s': 0.1, 'c':'k'},figsize=(20, 15))
>>> ax.set_xlim([0,200]); ax.set_ylim([0,200]); ax.set_zlim([0,200]);
getting atlas entry 1: image file /scratch/hpc3230/Data/conwhat_atlases/CWL2k8Sc33Vol3d100s_v01/vismap_grp_39-56_norm.nii.gz
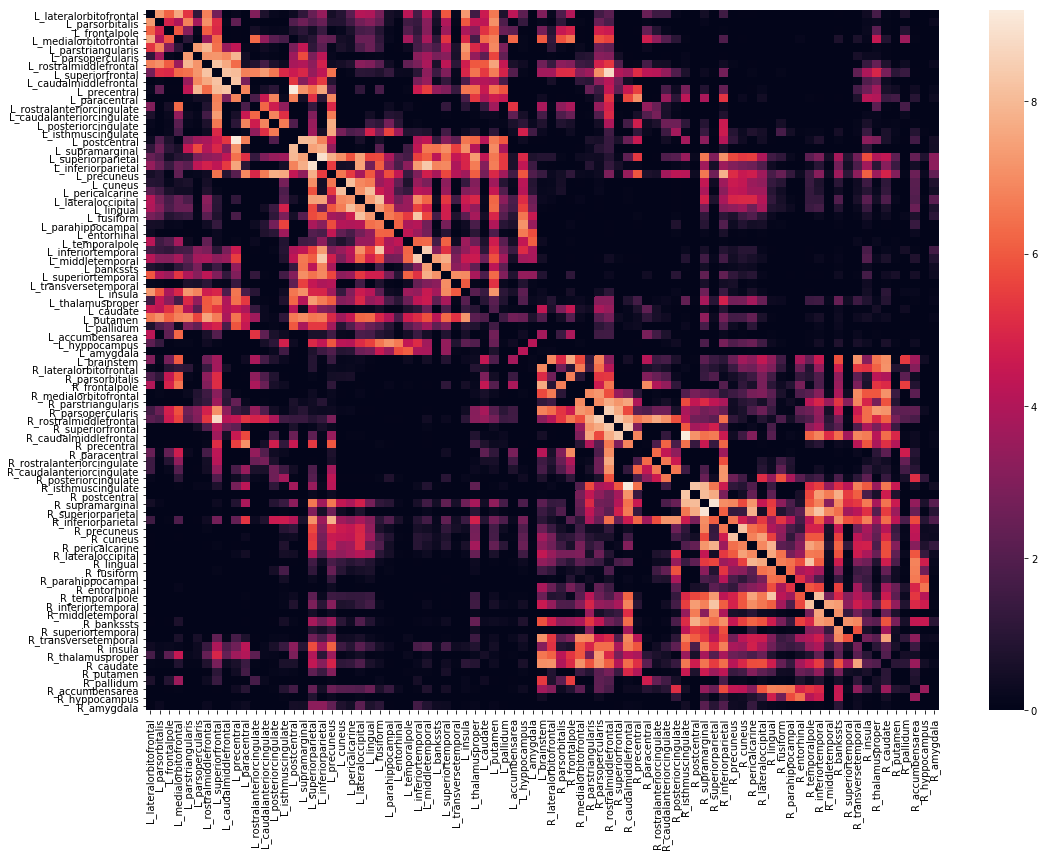
We can also view the weights matrix like so:
>>> fig, ax = plt.subplots(figsize=(16,12))
>>> sns.heatmap(np.log1p(vca.weights),xticklabels=vca.region_labels,
>>> yticklabels=vca.region_labels,ax=ax);
>>> plt.tight_layout()
The vca object also contains x,y,z bounding boxes for each structure
We also stored additional useful information about the ROIs in the associated parcellation, including cortical/subcortical labels
>>> vca.cortex
array([ 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 0., 0., 0., 0., 0.,
0., 0., 0., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 0., 0.,
0., 0., 0., 0., 0.])
…hemisphere labels
>>> vca.hemispheres
array([ 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1., 1.,
1., 1., 1., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0., 0.,
0., 0., 0., 0., 0.])
…and region mappings to freesurfer’s fsaverage brain
>>> vca.region_mapping_fsav_lh
array([ 24., 29., 28., ..., 16., 7., 7.])
>>> vca.region_mapping_fsav_rh
array([ 24., 29., 22., ..., 9., 9., 9.])
which can be used for, e.g. plotting ROI data on a surface
>>> f = '/opt/freesurfer/freesurfer/subjects/fsaverage/surf/lh.inflated'
>>> vtx,tri = nib.freesurfer.read_geometry(f)
>>> plot_surf_roi([vtx,tri],vca.region_mapping_fsav_lh);